Data Analysis Tools:
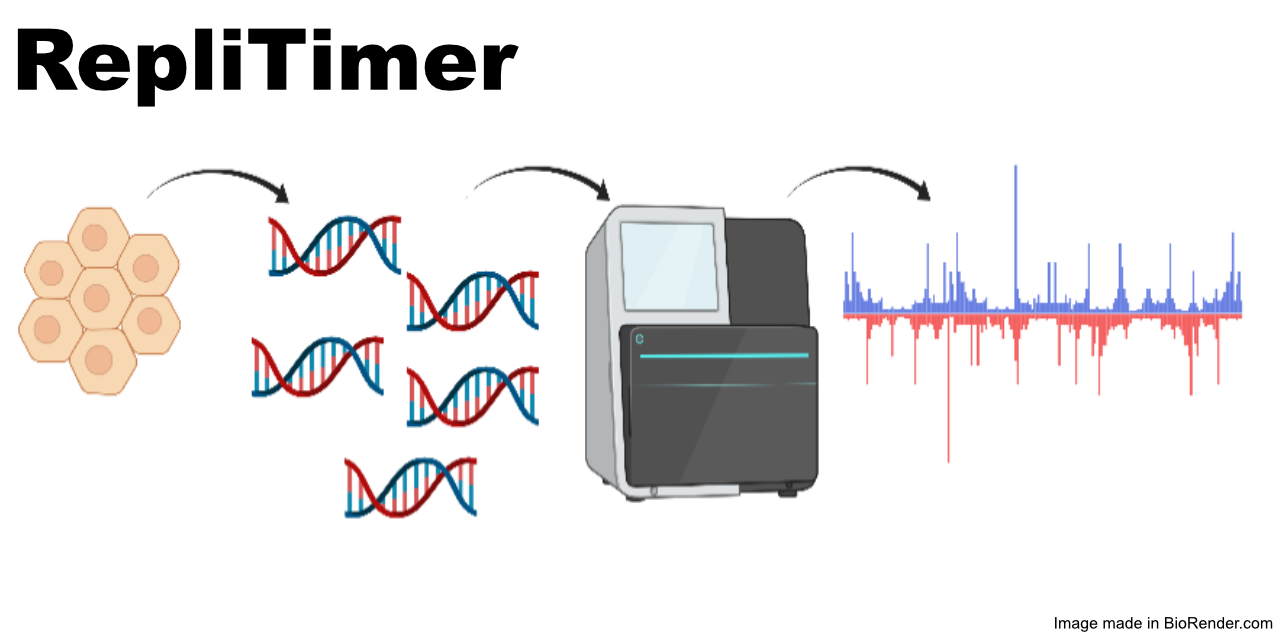
RepliTimer processes short whole-genome sequencing reads from any organism’s G1 and S phase cells into replication timing values. To enable step-by-step data processing, we describe each of the individual data processing steps. We provide a Snakemake pipeline with clearly defined dependencies and Anaconda environments to automate the pipeline. We include a compact dataset in the repository that you may use to test the pipeline. We also provide an example detailing how RepliTimer can be used to process publicly available replication timing data from zebrafish.
Cut&Run Analysis Snakemake
Cut_And_Run_Analysis_SnakeMake processes short whole-genome sequencing reads from Cut&Run. The pipeline generates trimmed fastq files, genome alignments, coverage files, and peak calls. To enable step-by-step data processing, we describe each of the individual data processing steps. We provide a Snakemake pipeline with clearly defined dependencies and Anaconda environments to automate the pipeline. We include a compact dataset in the repository that you may use to test the pipeline. We also provide an example detailing how Cut_And_Run_Analysis_SnakeMake can be used to process publicly available Cut&Run data. The original workflow was provided courtesy of the Sansam Lab.
QuantSeqPipeline
This Quantseq pipeline converts short read .fastq files from 3′ end sequencing into counts tables. The pipeline requires an index of the genome for the STAR aligner. After trimming to remove polyA sequences and adapters, the reads are aligned to the genome with STAR. Reads aligning to sequences adjacent to probable genomic internal polyA priming sites are removed. Reads are assigned to transcripts in a provided .gtf file and then a table a counts per transcript is generated. This table can be used for statistical analysis using DESeq2 or other statistical analysis software. This pipeline is comprised of shell scripts that can be executed on SLURM using the provided wrapper script. Conda environments are not provided, so please consult with your HPC administrator to run this.
CRISPR Oligo Maker









 Find Christopher Sansam Lab Plasmids
Find Christopher Sansam Lab Plasmids